|
|
|
|
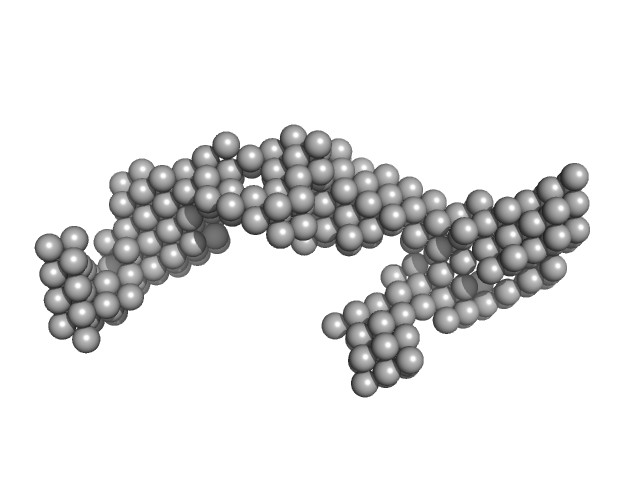
|
| Sample: |
DENV2 3' untranslated region monomer, 160 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 10
|
Structural Dynamics of Dengue Virus UTRs and Their Cyclization.
Biophys J (2025)
Robinson ZE, Pereira HS, D'Souza MH, Patel TR
|
| RgGuinier |
6.0 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|
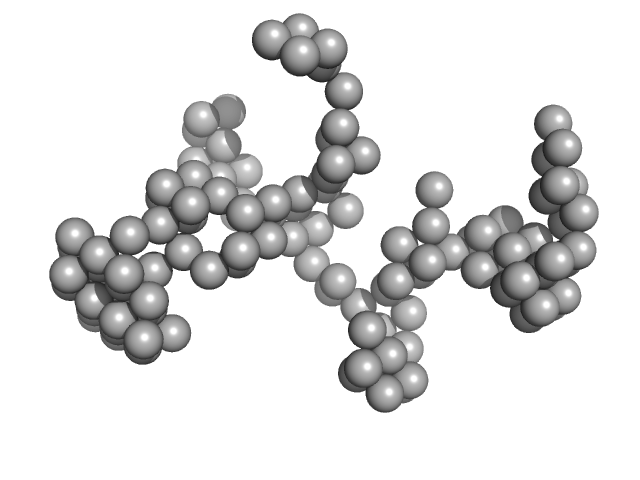
|
| Sample: |
DENV2 complex 5'-3' monomer, 217 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 10
|
Structural Dynamics of Dengue Virus UTRs and Their Cyclization.
Biophys J (2025)
Robinson ZE, Pereira HS, D'Souza MH, Patel TR
|
| RgGuinier |
7.5 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
320 |
nm3 |
|
|
|
|
|
|
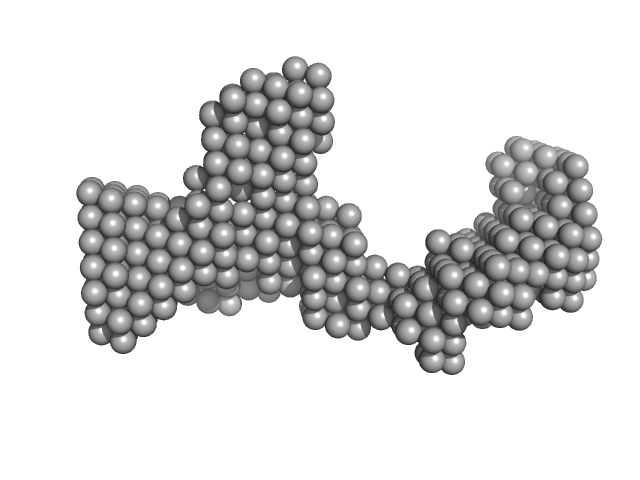
|
| Sample: |
DENV2 RNA, 5' Untranslated region monomer, 57 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 8
|
Structural Dynamics of Dengue Virus UTRs and Their Cyclization.
Biophys J (2025)
Robinson ZE, Pereira HS, D'Souza MH, Patel TR
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
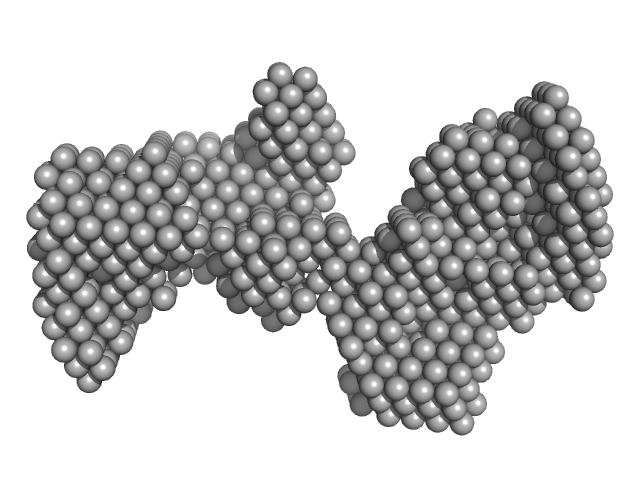
|
| Sample: |
G4 dumbbell RNA - 3' untranslated region (3' UTR) Dengue virus monomer, 31 kDa dengue virus type … RNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 0.01 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 9
|
Unravelling structural dynamics of Dengue virus 3' UTR and its impact on viral life cycle
Trushar Patel
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dumbbell RNA - 3' untranslated region (3' UTR) Dengue virus monomer, 31 kDa dengue virus type … RNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 9
|
Unravelling structural dynamics of Dengue virus 3' UTR and its impact on viral life cycle
Trushar Patel
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
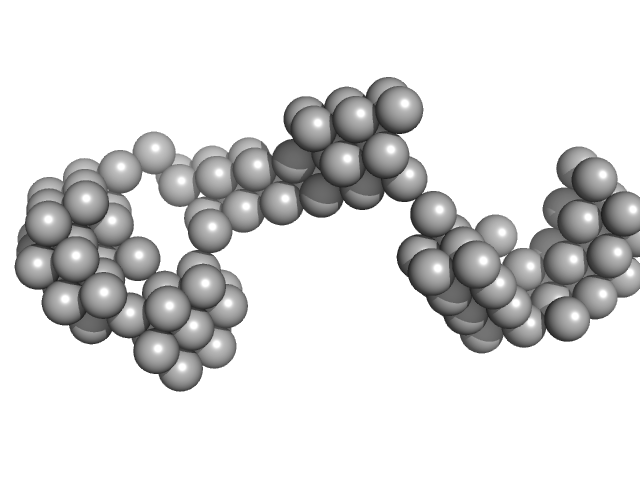
| Sample: |
G4 dumbbell Mut - 3' untranslated region (3' UTR) Dengue virus dimer, 61 kDa dengue virus type … RNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 0.01 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jun 30
|
Unravelling structural dynamics of Dengue virus 3' UTR and its impact on viral life cycle
Trushar Patel
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GAGE6 60bp dsDNA oligo monomer, 37 kDa Homo sapiens DNA
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 14
|
Probing the statistics of sequence-dependent DNA conformations in solution using SAXS
Heidar Koning
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
|
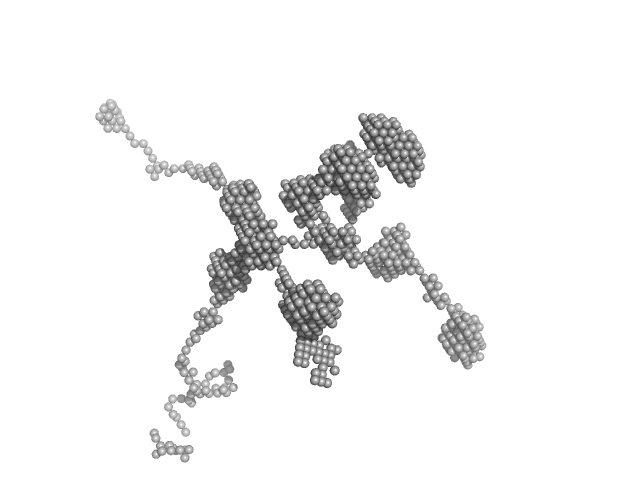
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA vaccines
Nature Nanotechnology (2025)
Moreno Herrero J, Stahl T, Erbar S, Maxeiner K, Schlegel A, Bacic T, Schumacher J, Cavalcanti L, Schroer M, Svergun D, Sahin U, Haas H
|
| RgGuinier |
90.5 |
nm |
| Dmax |
200.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 10
|
Compact polyethylenimine-complexed mRNA vaccines
Nature Nanotechnology (2025)
Moreno Herrero J, Stahl T, Erbar S, Maxeiner K, Schlegel A, Bacic T, Schumacher J, Cavalcanti L, Schroer M, Svergun D, Sahin U, Haas H
|
| RgGuinier |
30.7 |
nm |
| Dmax |
107.4 |
nm |
| VolumePorod |
112000 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
Linear polyethylenimine monomer, 25 kDa none (polymer)
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA vaccines
Nature Nanotechnology (2025)
Moreno Herrero J, Stahl T, Erbar S, Maxeiner K, Schlegel A, Bacic T, Schumacher J, Cavalcanti L, Schroer M, Svergun D, Sahin U, Haas H
|
| RgGuinier |
12.0 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
9100 |
nm3 |
|
|