|
|
|
|
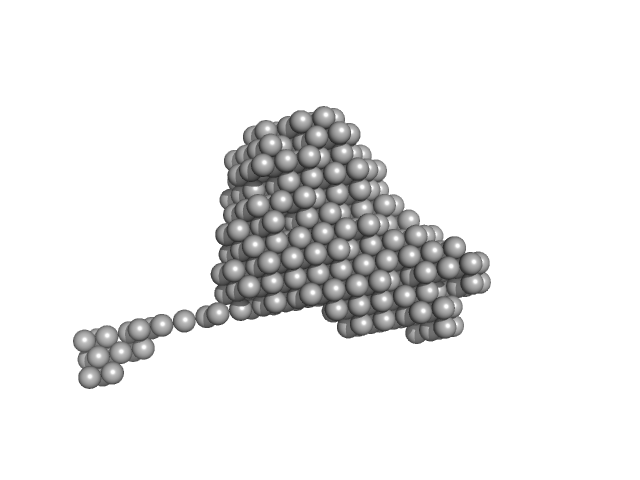
|
| Sample: |
Endonuclease 8-like 1 monomer, 45 kDa Homo sapiens protein
DsDNA monomer, 2 kDa DNA
Proliferating cell nuclear antigen monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
25mM HEPES 100mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Jan 20
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
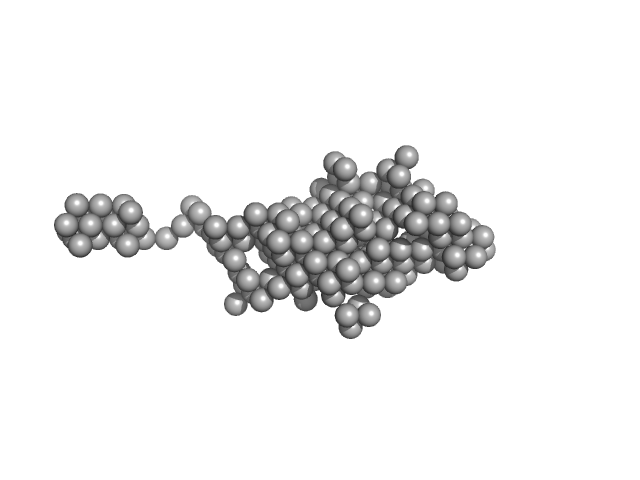
|
| Sample: |
Endonuclease 8-like 1 monomer, 45 kDa Homo sapiens protein
DsDNA monomer, 2 kDa DNA
|
| Buffer: |
25mM HEPES 100mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Jan 20
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
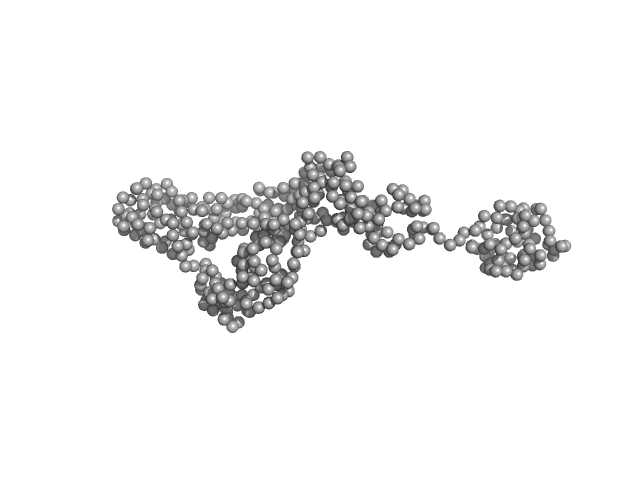
|
| Sample: |
Endonuclease 8-like 1 monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
25mM HEPES 300mM NaCl 1mM DTT 10% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 13
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
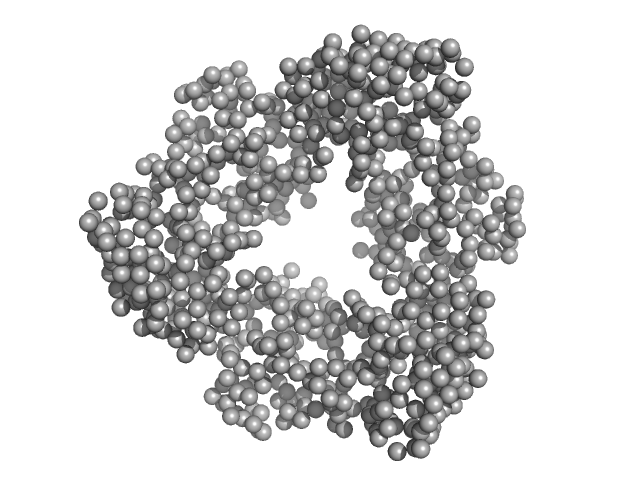
|
| Sample: |
Proliferating cell nuclear antigen trimer, 89 kDa Homo sapiens protein
|
| Buffer: |
25mM HEPES 100mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Jan 20
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
|
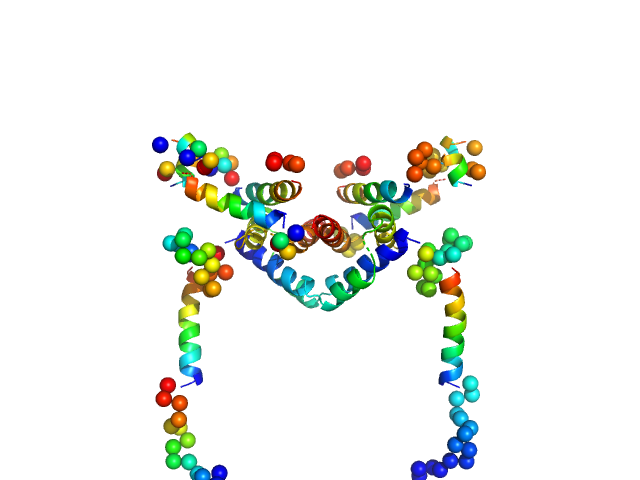
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|
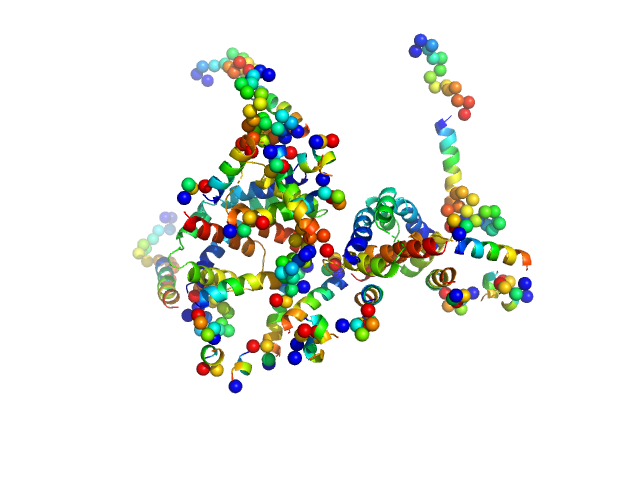
|
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) tetramer, 85 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-crystallin B2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM NaPi, 5 mM DTT, 1 mM EDTA,, pH: 6.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 20
|
Human βB2-Crystallin Forms a Face-en-Face Dimer in Solution: An Integrated NMR and SAXS Study.
Structure 25(3):496-505 (2017)
Xi Z, Whitley MJ, Gronenborn AM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-crystallin B2 dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
25 mM NaPi, 5 mM DTT, 1 mM EDTA,, pH: 6.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 20
|
Human βB2-Crystallin Forms a Face-en-Face Dimer in Solution: An Integrated NMR and SAXS Study.
Structure 25(3):496-505 (2017)
Xi Z, Whitley MJ, Gronenborn AM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
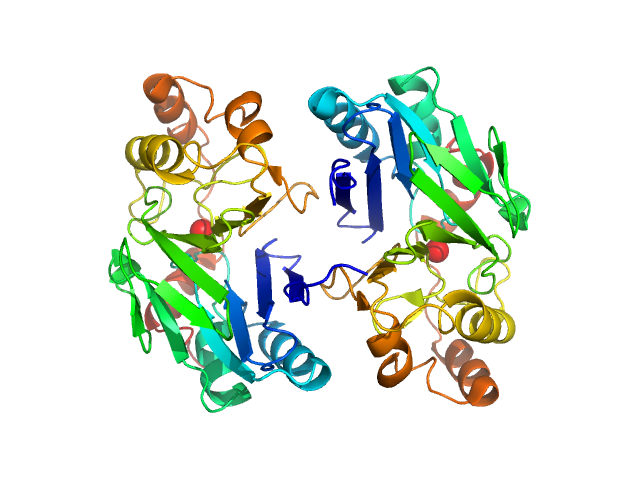
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 21
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
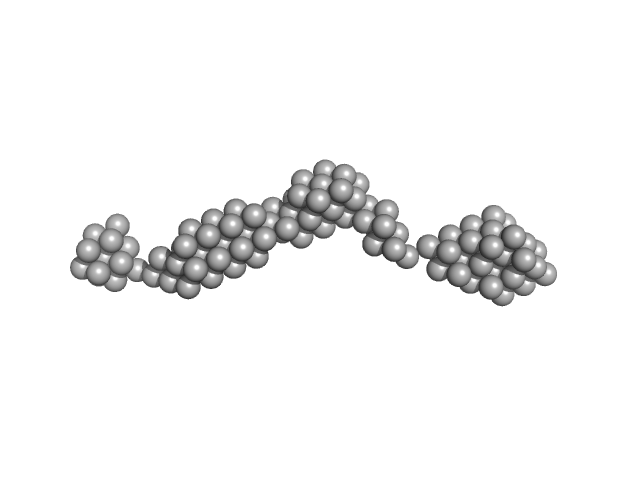
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
9.0 |
nm |
| Dmax |
32.3 |
nm |
| VolumePorod |
400 |
nm3 |
|
|