|
|
|
|
|
| Sample: |
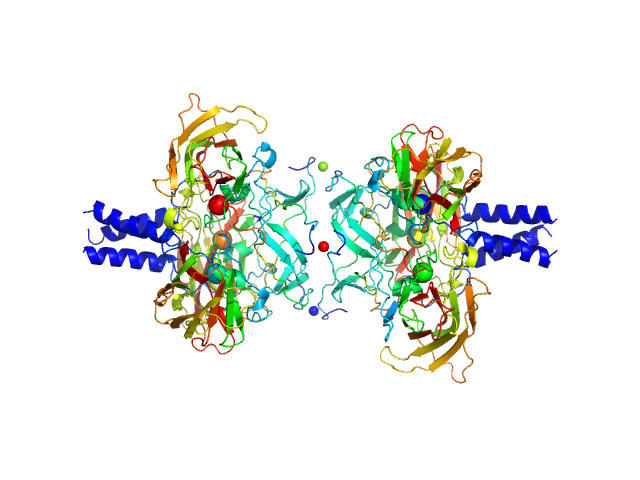
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
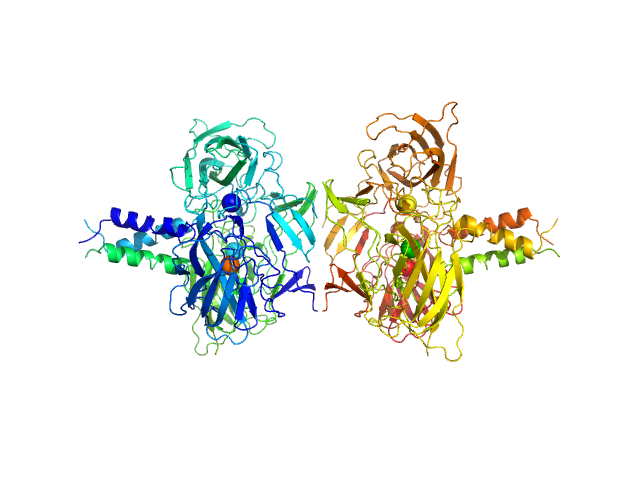
SUN domain-containing protein 1 hexamer, 135 kDa Homo sapiens protein
Nesprin-4 hexamer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 21
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
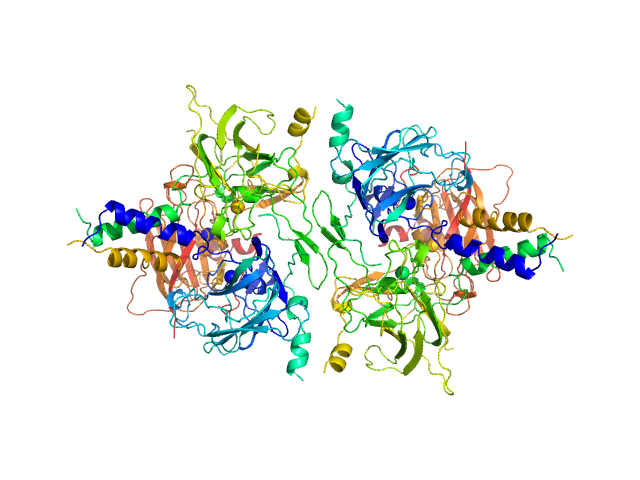
SUN domain-containing protein 1 hexamer, 135 kDa Homo sapiens protein
Protein KASH5 hexamer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 17
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
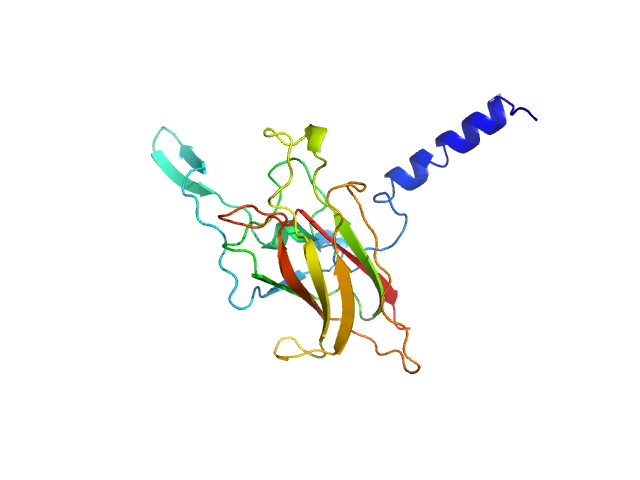
SUN domain-containing protein 1 hexamer, 135 kDa Homo sapiens protein
Nesprin-1 hexamer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 17
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SUN domain-containing protein 1, I673E monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jul 29
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
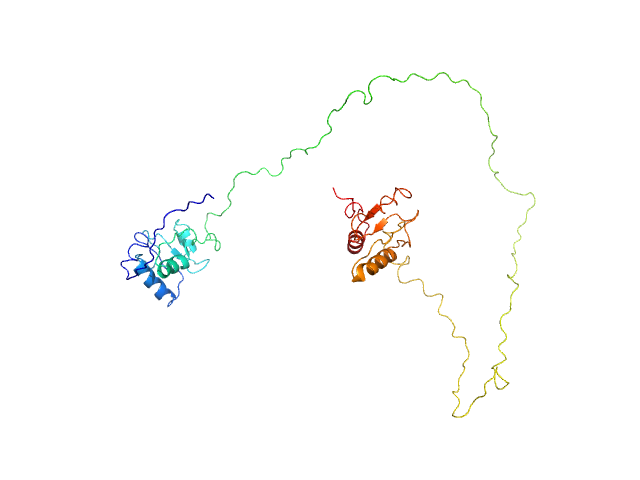
| Sample: |
DNA repair protein XRCC1ΔN dimer, 76 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Jul 15
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
|
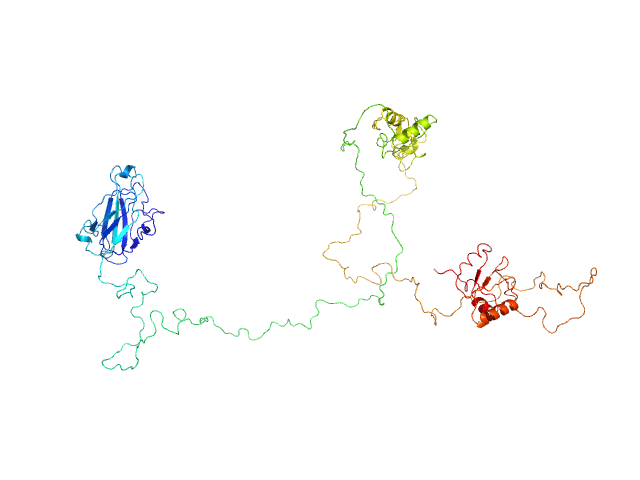
| Sample: |
DNA repair protein XRCC1, 69 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 19
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.3 |
nm |
| Dmax |
26.9 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
|
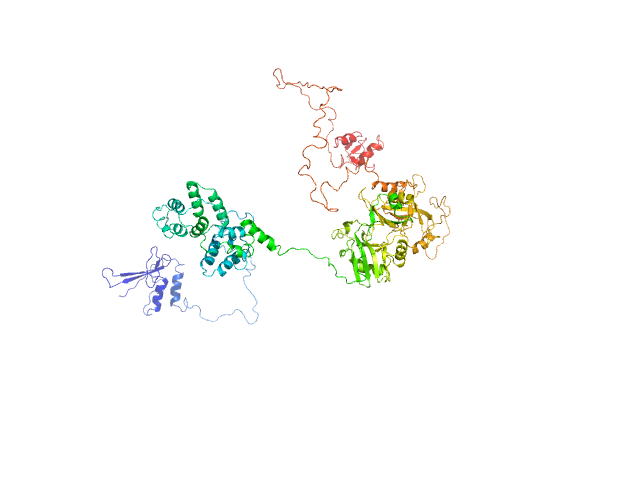
| Sample: |
DNA ligase 3 (DNA ligase III alpha), Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Mar 31
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
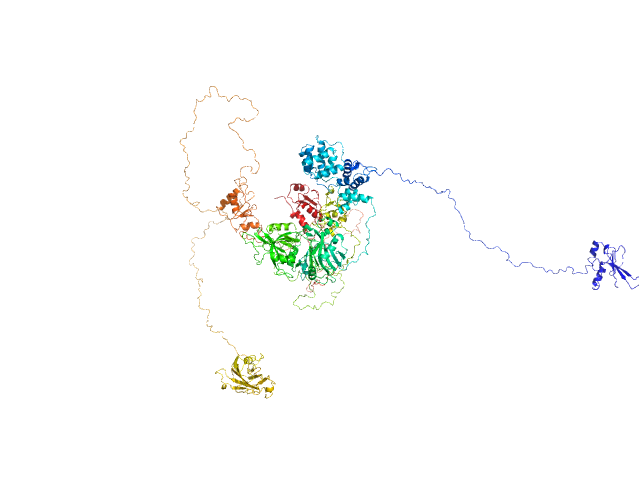
| Sample: |
DNA repair protein XRCC1, 69 kDa Homo sapiens protein
DNA ligase 3 (DNA ligase III alpha), Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl₂, 0.2 mM PMSF, 1 mM benzamidine, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Aug 18
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.2 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
675 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 7
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
117 |
nm3 |
|
|