|
|
|
|
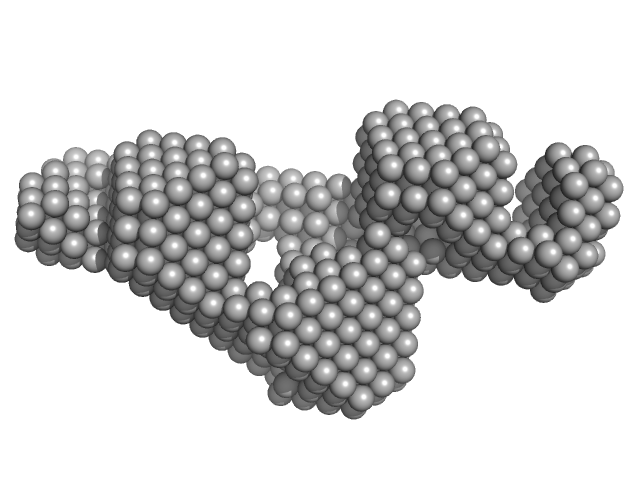
|
| Sample: |
Grp1 63-399 E161A 6GS Arf6 Q67L SUMO fusion protein monomer, 72 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Grp1 63-399 E161A Arf6 Q67L fusion protein monomer, 60 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sortilin 1 A464E alias Neurotensin-receptor 3 A464E monomer, 76 kDa Mus musculus protein
|
| Buffer: |
25 mM MES pH 5.5, 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
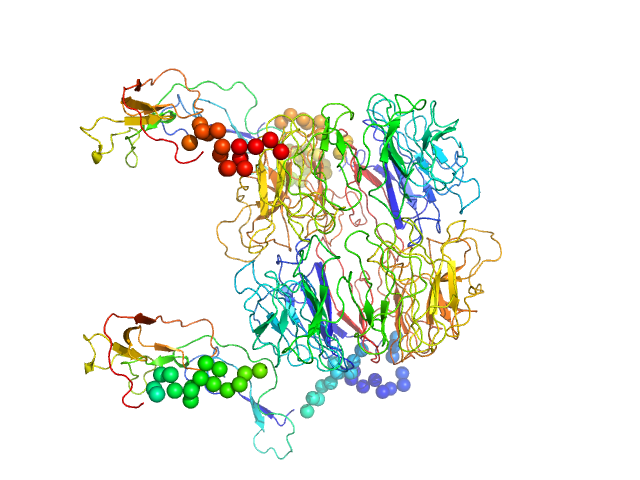
|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM MES pH 5.5, 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
336 |
nm3 |
|
|
|
|
|
|
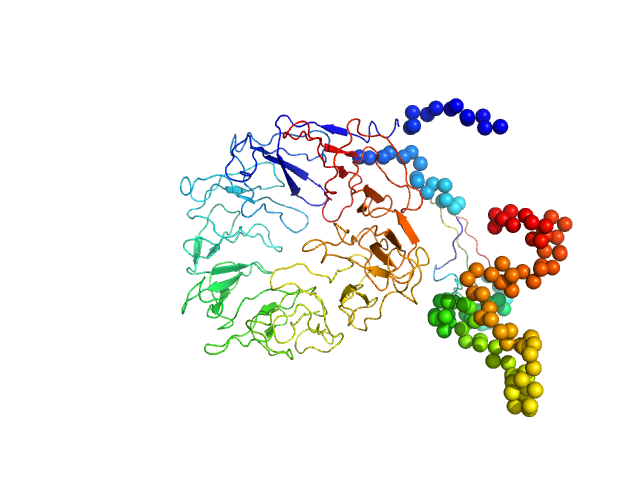
|
| Sample: |
Sortilin 1 A464E alias Neurotensin-receptor 3 A464E monomer, 76 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
|
|
|
|
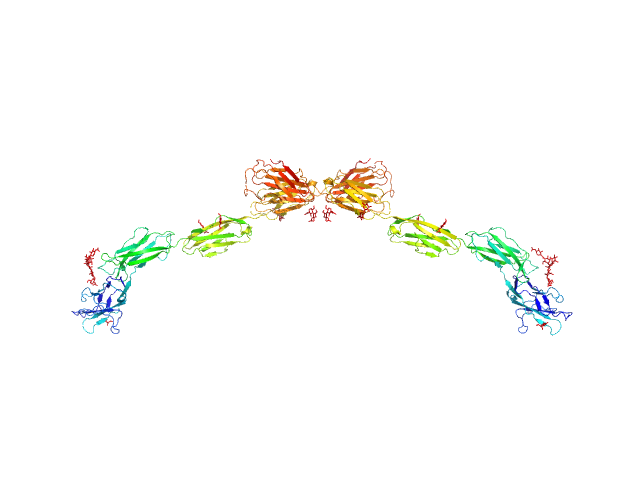
|
| Sample: |
Myelin-associated glycoprotein Ig domains 1-5 dimer, 108 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.8 |
nm |
| Dmax |
23.8 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
|
|
|
|
|
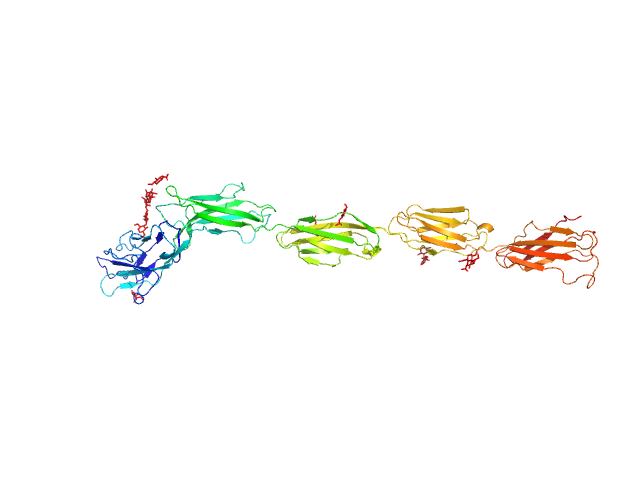
| Sample: |
Myelin-associated glycoprotein (20-508; I473E mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
117 |
nm3 |
|
|