|
|
|
|
|
| Sample: |
Major viral transcription factor ICP4 dimer, 49 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jul 27
|
The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res 45(13):8064-8078 (2017)
Tunnicliffe RB, Lockhart-Cairns MP, Levy C, Mould AP, Jowitt TA, Sito H, Baldock C, Sandri-Goldin RM, Golovanov AP
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
Immediate Early 3 dimer, 12 kDa DNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Nov 12
|
The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res 45(13):8064-8078 (2017)
Tunnicliffe RB, Lockhart-Cairns MP, Levy C, Mould AP, Jowitt TA, Sito H, Baldock C, Sandri-Goldin RM, Golovanov AP
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
C-terminal catalytic domain of Suppressor of Copper Sensitivity C protein monomer, 20 kDa Proteus mirabilis protein
|
| Buffer: |
25 mM HEPES 150mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2012 Feb 29
|
A shape-shifting redox foldase contributes to Proteus mirabilis copper resistance.
Nat Commun 8:16065 (2017)
Furlong EJ, Lo AW, Kurth F, Premkumar L, Totsika M, Achard MES, Halili MA, Heras B, Whitten AE, Choudhury HG, Schembri MA, Martin JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Suppressor of Copper Sensitivity C protein (mutant) trimer, 73 kDa Proteus mirabilis protein
|
| Buffer: |
25 mM HEPES 150mM NaCl, 1mM DTT,, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Jul 22
|
A shape-shifting redox foldase contributes to Proteus mirabilis copper resistance.
Nat Commun 8:16065 (2017)
Furlong EJ, Lo AW, Kurth F, Premkumar L, Totsika M, Achard MES, Halili MA, Heras B, Whitten AE, Choudhury HG, Schembri MA, Martin JL
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
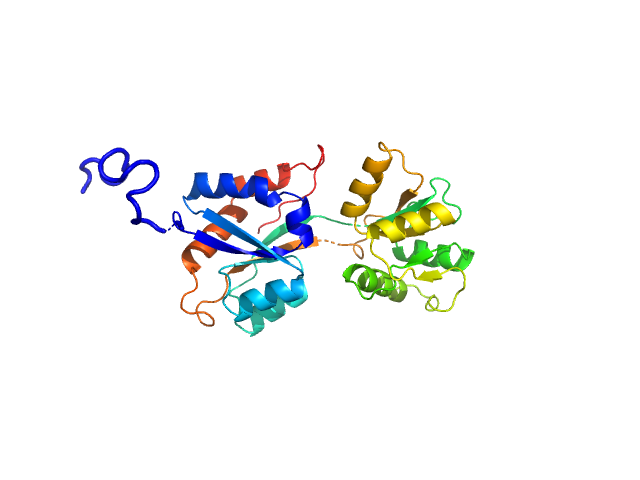
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
|
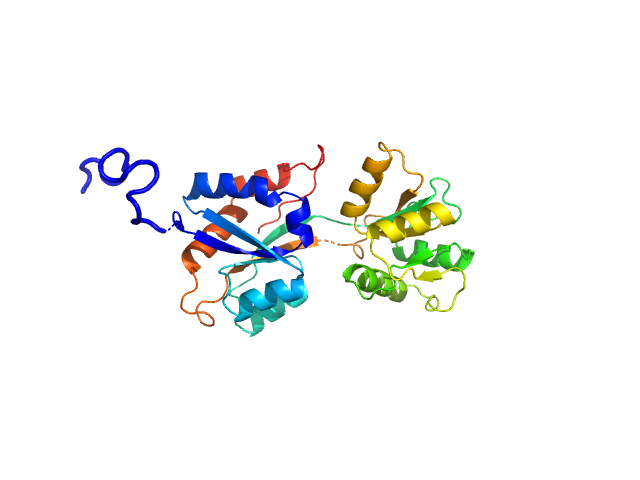
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
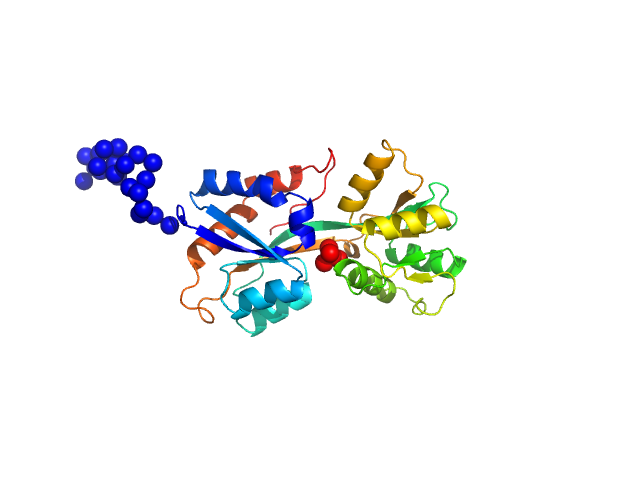
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
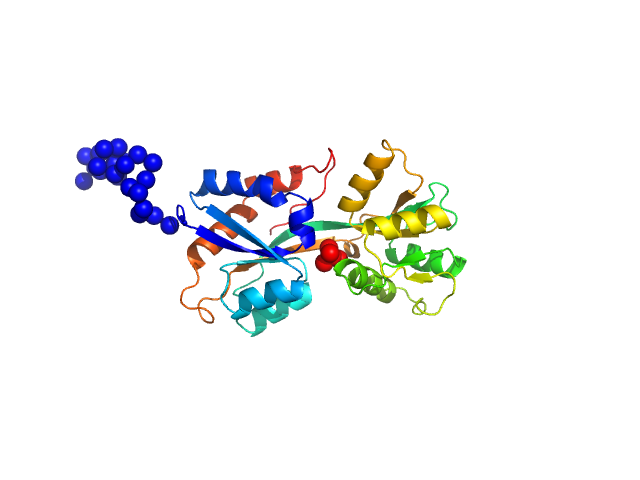
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
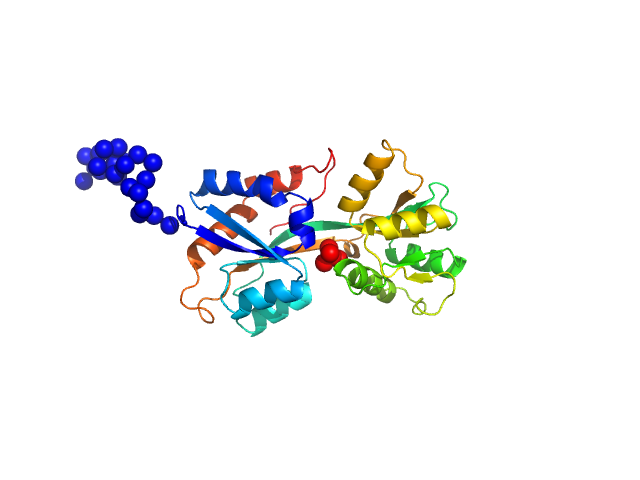
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
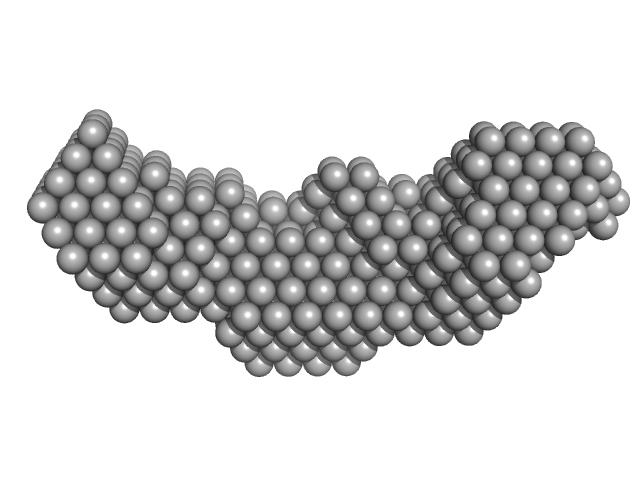
|
| Sample: |
Homeobox protein CEH-14 monomer, 16 kDa Caenorhabditis elegans protein
CeLIM-7 monomer, 4 kDa Caenorhabditis elegans protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 5 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Sydney on 2009 Apr 7
|
Interactions between LHX3- and ISL1-family LIM-homeodomain transcription factors are conserved in Caenorhabditis elegans.
Sci Rep 7(1):4579 (2017)
Bhati M, Llamosas E, Jacques DA, Jeffries CM, Dastmalchi S, Ripin N, Nicholas HR, Matthews JM
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
26 |
nm3 |
|
|