|
|
|
|
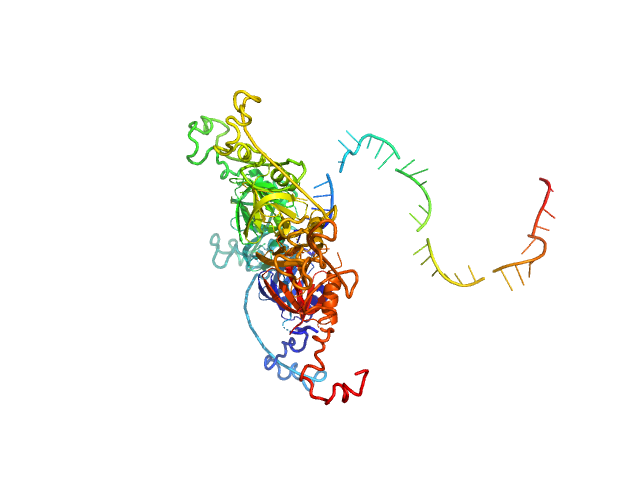
|
| Sample: |
RNA chaperone Hfq hexamer, 67 kDa Escherichia coli protein
RNA DsrA monomer, 12 kDa RNA
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 16
|
Structural flexibility of RNA as molecular basis for Hfq chaperone function.
Nucleic Acids Res 40(16):8072-84 (2012)
...Konarev PV, Shang W, Vecerek B, Kontaxis G, Hämmerle H, Peterlik H, Svergun DI, Bläsi U, Djinović-Carugo K
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
210 |
nm3 |
|