|
|
|
|
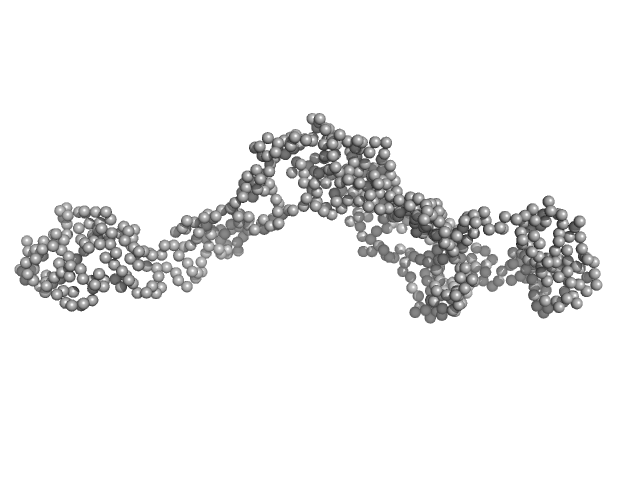
|
| Sample: |
Tyrosine-protein kinase BTK (R28C mutant) monomer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, 2 mM DTT and 1 mM MgCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Apr 2
|
Conformation of full-length Bruton tyrosine kinase (Btk) from synchrotron X-ray solution scattering.
EMBO J 22(18):4616-24 (2003)
...Smith CI, Petoukhov MV, Lo Surdo P, Mattsson PT, Knekt M, Westlund A, Scheffzek K, Saraste M, Svergun DI
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
130 |
nm3 |
|