UniProt ID: Q9AES2 (323-878) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 64 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Apr 19
|
Molecular basis of Fab-dependent IgA antibody recognition by gut-bacterial metallopeptidases
The EMBO Journal (2025)
Márquez-Moñino M, Martínez Gascueña A, Azzam T, Persson A, Manzanares-Gomez A, Aguillo-Urarte M, Brown T, Montero-Sagarminaga A, Lood R, Naegeli A, Connell S, Sastre D, Sundberg E, Trastoy B
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: A0AAE9RTQ7 (43-265) Virulence factor family protein
|
|
|
|
| Sample: |
Virulence factor family protein monomer, 24 kDa Brucella abortus protein
|
| Buffer: |
50 mM Tris pH 8.0, 200 mM NaCl, 5% v/v glycerol, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Jul 12
|
Crystal structure of the virulence protein J (VirJ) domain 1 from Brucella abortus.
Acta Crystallogr F Struct Biol Commun (2025)
Dugelay C, Ferrarin S, Terradot L
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: A0A0D1JF17 (1-109) Cyclic-di-AMP receptor
|
|
|
|
| Sample: |
Cyclic-di-AMP receptor trimer, 42 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 11
|
Activation of a Secondary-Messenger Receptor via Allosteric Modulation of a Dynamic Conformational Ensemble.
Angew Chem Int Ed Engl :e202509394 (2025)
Söldner B, Singh H, Akoury E, Witte G, Linser R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
UniProt ID: A0A0D1JF17 (1-109) Cyclic-di-AMP receptor
|
|
|
|
| Sample: |
Cyclic-di-AMP receptor trimer, 42 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 11
|
Activation of a Secondary-Messenger Receptor via Allosteric Modulation of a Dynamic Conformational Ensemble.
Angew Chem Int Ed Engl :e202509394 (2025)
Söldner B, Singh H, Akoury E, Witte G, Linser R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
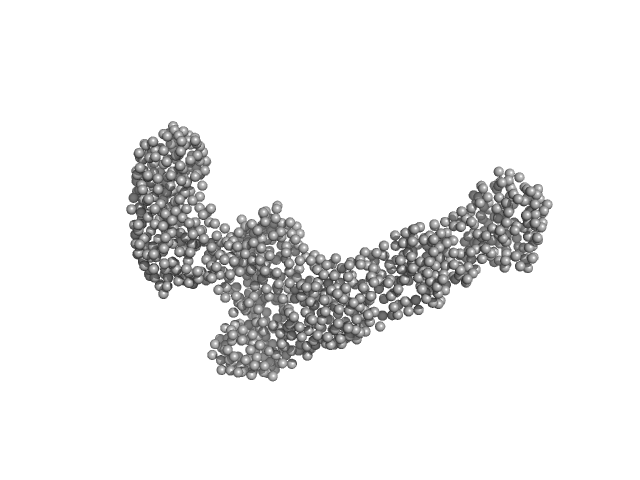
UniProt ID: P0A6Y8 (None-None) Chaperone protein DnaK
UniProt ID: Q8IL04 (None-None) Heme ligase
|
|
|
|
| Sample: |
Chaperone protein DnaK, 69 kDa Escherichia coli (strain … protein
Heme ligase, 28 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, Chemistry Division, Bhabha Atomic Research Centre on 2019 Feb 26
|
Recombinant expression and purification of Plasmodium heme detoxification protein in E. coli: Challenges and discoveries.
Protein Expr Purif 236:106789 (2025)
Singh R, Makde RD
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
UniProt ID: Q9UQM7 (7-478) Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344)
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344) dodecamer, 620 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris , 0.2 mM CaCl2, 150 mM KCl, 10 mM MgCl2, 2 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku/BioSAXS-2000 SAXS, Institute for Applied Life Sciences/Biophysical Characterization on 2024 Dec 18
|
A domain-swapped CaMKII conformation facilitates linker-mediated allosteric regulation - revision
Bao Nguyen Viet
|
|
|
UniProt ID: Q9UQM7 (7-478) Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344)
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344) dodecamer, 620 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris , 0.2 mM CaCl2, 150 mM KCl, 10 mM MgCl2, 2 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku/BioSAXS-2000 SAXS, Institute for Applied Life Sciences/Biophysical Characterization on 2024 Dec 18
|
A domain-swapped CaMKII conformation facilitates linker-mediated allosteric regulation - revision
Bao Nguyen Viet
|
|
|
UniProt ID: Q9UQM7 (7-478) Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344)
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344) dodecamer, 620 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris , 0.2 mM CaCl2, 150 mM KCl, 10 mM MgCl2, 2 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku/BioSAXS-2000 SAXS, Institute for Applied Life Sciences/Biophysical Characterization on 2024 Dec 18
|
A domain-swapped CaMKII conformation facilitates linker-mediated allosteric regulation - revision
Bao Nguyen Viet
|
|
|
UniProt ID: Q9UQM7 (7-478) Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344)
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344) dodecamer, 620 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris , 0.2 mM CaCl2, 150 mM KCl, 10 mM MgCl2, 2 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku/BioSAXS-2000 SAXS, Institute for Applied Life Sciences/Biophysical Characterization on 2024 Dec 18
|
A domain-swapped CaMKII conformation facilitates linker-mediated allosteric regulation - revision
Bao Nguyen Viet
|
|
|
UniProt ID: Q9UQM7 (7-478) Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ 316-329)
|
|
|
|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ 316-329) dodecamer, 626 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris , 0.2 mM CaCl2, 150 mM KCl, 10 mM MgCl2, 2 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku/BioSAXS-2000 SAXS, Institute for Applied Life Sciences/Biophysical Characterization on 2024 Dec 18
|
A domain-swapped CaMKII conformation facilitates linker-mediated allosteric regulation - revision
Bao Nguyen Viet
|
|
|