UniProt ID: P42196 (1-239) Sensory rhodopsin II from Natronbacterium pharaonis
UniProt ID: P42259 (3-534) Sensory rhodopsin II transducer from Natronomonas pharaonis
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 9
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
9.0 |
nm |
| Dmax |
38.9 |
nm |
|
|
UniProt ID: P25440 (71-455) Bromodomain-containing protein 2
|
|
|
|
| Sample: |
Bromodomain-containing protein 2 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 25
|
Multivalent nucleosome scaffolding by bromodomain and extraterminal domain tandem bromodomains.
J Biol Chem :108289 (2025)
Olp MD, Bursch KL, Wynia-Smith SL, Nuñez R, Goetz CJ, Jackson V, Smith BC
|
| RgGuinier |
4.9 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
UniProt ID: Q7DD94 (1-544) YhbX/YhjW/YijP/YjdB family protein (L152F)
|
|
|
|
| Sample: |
YhbX/YhjW/YijP/YjdB family protein (L152F) monomer, 62 kDa Neisseria meningitidis serogroup … protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 0.14% Fos-Choline 12 (FC-12), pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 9
|
Conformational flexibility of EptA driven by an interdomain helix provides insights for enzyme-substrate recognition.
IUCrJ 8(Pt 5):732-746 (2021)
Anandan A, Dunstan NW, Ryan TM, Mertens HDT, Lim KYL, Evans GL, Kahler CM, Vrielink A
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
UniProt ID: B7UM99 (60-200) Translocated intimin receptor Tir
|
|
|
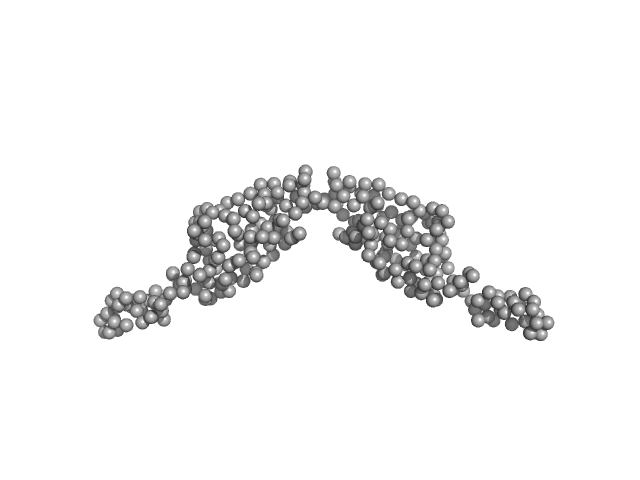
|
| Sample: |
Translocated intimin receptor Tir dimer, 32 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: P23615 (None-None) Transcription elongation factor SPT6 - ΔtSH2 variant
|
|
|
|
| Sample: |
Transcription elongation factor SPT6 - ΔtSH2 variant monomer, 122 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
25 mM NaPi; 150mM NaCl; 0.5 mM EDTA; 5% glycerol; 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 22
|
Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res (2022)
Kasiliauskaite A, Kubicek K, Klumpler T, Zanova M, Zapletal D, Koutna E, Novacek J, Stefl R
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
245 |
nm3 |
|
|
UniProt ID: E0IW31 (90-1520) Accessory colonization factor
|
|
|

|
| Sample: |
Accessory colonization factor monomer, 160 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM citrate-phosphate buffer, 200 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 27
|
Molecular and cellular insight into Escherichia coli SslE and its role during biofilm maturation
npj Biofilms and Microbiomes 8(1) (2022)
Corsini P, Wang S, Rehman S, Fenn K, Sagar A, Sirovica S, Cleaver L, Edwards-Gayle C, Mastroianni G, Dorgan B, Sewell L, Lynham S, Iuga D, Franks W, Jarvis J, Carpenter G, Curtis M, Bernadó P, Darbari V, Garnett J
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
UniProt ID: Q9GZZ9 (57-346) Ubiquitin-like modifier-activating enzyme 5
UniProt ID: P61960 (1-83) Ubiquitin fold modifer 1
|
|
|
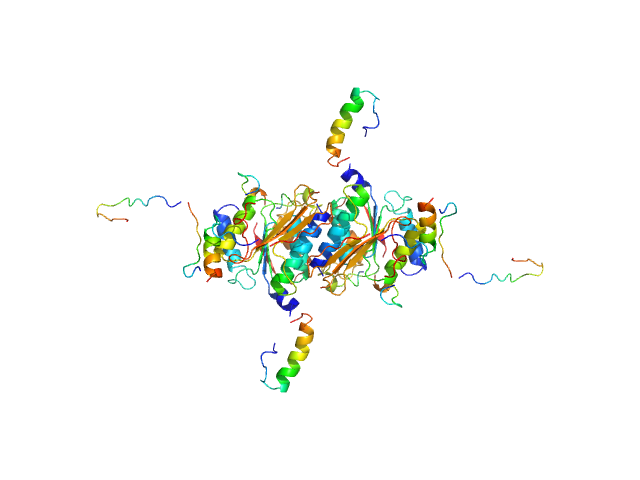
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
|
|
UniProt ID: P20809 (32-199) Interleukin-11
UniProt ID: Q14626 (23-319) Interleukin-11 receptor subunit alpha
UniProt ID: P40189 (23-612) Interleukin-6 receptor subunit beta
|
|
|
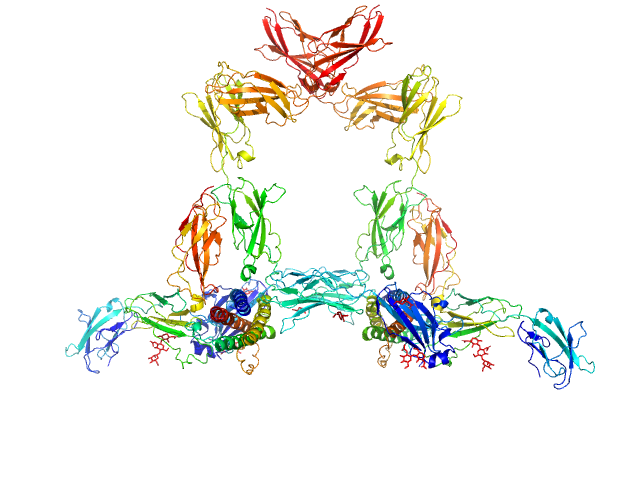
|
| Sample: |
Interleukin-11 dimer, 36 kDa Homo sapiens protein
Interleukin-11 receptor subunit alpha dimer, 64 kDa Homo sapiens protein
Interleukin-6 receptor subunit beta dimer, 134 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 28
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
710 |
nm3 |
|
|
UniProt ID: P54253 (562-815) Ataxin-1
|
|
|
|
| Sample: |
Ataxin-1 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
|
|