|
|
|
|
|
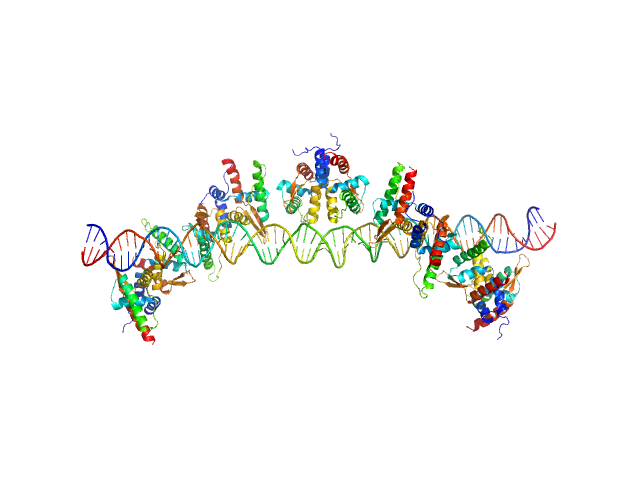
| Sample: |
GraTA operator region monomer, 20 kDa DNA
Antitoxin GraA, antidote of toxin GraT dimer, 22 kDa Pseudomonas putida protein
Atitoxin GraA, antidote of GraT. dimer, 23 kDa Pseudomonas putida protein
|
| Buffer: |
50 mM Tris 250 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Dec 17
|
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun 10(1):972 (2019)
Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
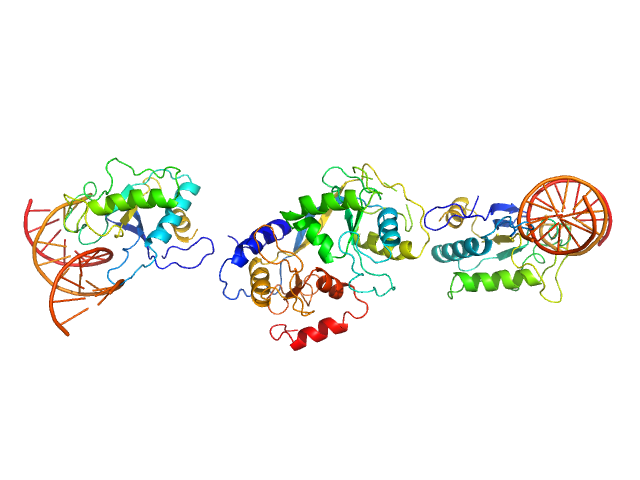
| Sample: |
S48 DNA strand 1 monomer, 21 kDa DNA
S48 DNA strand 2 monomer, 21 kDa DNA
TubR of the pXO1-like plasmid pBc10987 from B. cereus (Bc-TubR) decamer, 137 kDa protein
|
| Buffer: |
0.1 M NaCl, 10 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Nov 28
|
Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.
J Mol Biol (2018)
Hayashi I, Oda T, Sato M, Fuchigami S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
305 |
nm3 |
|
|
|
|
|
|
|
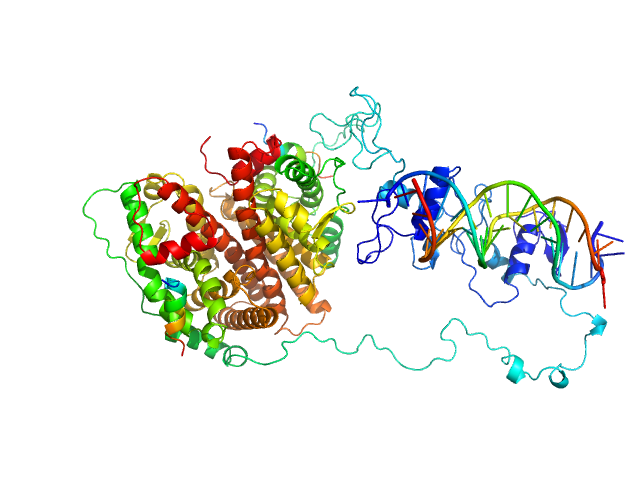
| Sample: |
Cognate hemimethylated 12-bp oligoduplex dimer, 15 kDa DNA
5-methylcytosine-specific restriction enzyme A dimer, 65 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
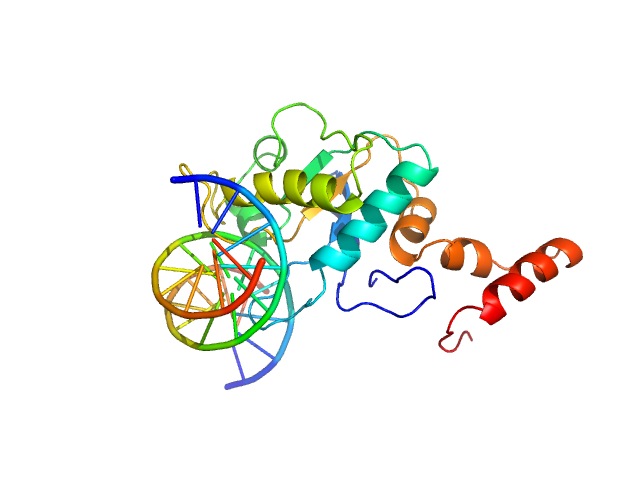
|
| Sample: |
5-methylcytosine-specific restriction enzyme A (N-terminal domain) monomer, 21 kDa Escherichia coli protein
Cognate hemimethylated 12-bp oligoduplex monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor dimer, 85 kDa protein
ERE1 monomer, 6 kDa Homo sapiens DNA
ERE2 monomer, 6 kDa Homo sapiens DNA
Estradiol dimer, 0 kDa
HERa peptide1 monomer, 2 kDa protein
HERa peptide2 monomer, 2 kDa protein
|
| Buffer: |
10 mM CHES (pH9.5), 125 mM NaCl, 5mM KCl, 4 mM MgCl2, 50 mM arginine, 50 mM glutamate, 5 mM TCEP, 5% glycerol, 10 µm Zn acetate, 10 µM estradiol, pH: 9.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Aug 10
|
Multidomain architecture of estrogen receptor reveals interfacial cross-talk between its DNA-binding and ligand-binding domains.
Nat Commun 9(1):3520 (2018)
Huang W, Peng Y, Kiselar J, Zhao X, Albaqami A, Mendez D, Chen Y, Chakravarthy S, Gupta S, Ralston C, Kao HY, Chance MR, Yang S
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
12.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, 0.5 mM AMP-PNP, pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.6 |
nm |
| Dmax |
16.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
Nonstructural protein sigma NS octamer, 325 kDa Avian orthoreovirus protein
20mer RNA (unstructured) dimer, 13 kDa RNA
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
Stability of local secondary structure determines selectivity of viral RNA chaperones.
Nucleic Acids Res (2018)
Bravo JPK, Borodavka A, Barth A, Calabrese AN, Mojzes P, Cockburn JJB, Lamb DC, Tuma R
|
| RgGuinier |
7.8 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
964 |
nm3 |
|
|