|
|
|
|
|
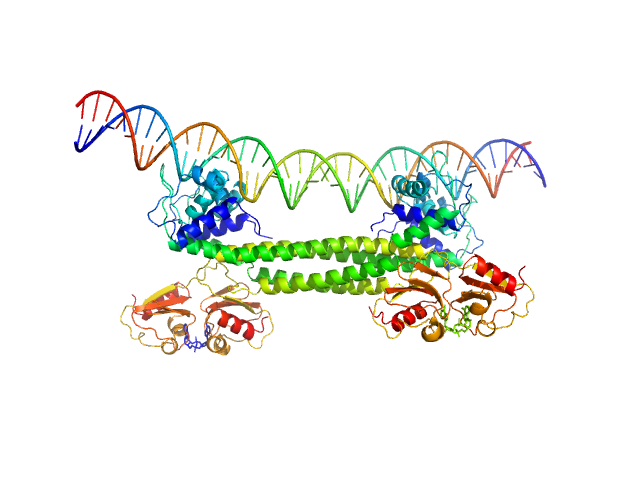
| Sample: |
Transcriptional repressor BusR tetramer, 95 kDa Streptococcus agalactiae protein
BusR Recognition sequence monomer, 28 kDa synthetic construct DNA
|
| Buffer: |
20mM HEPES, pH6.5, 100mM NaCl, 3% glycerol (v/v), pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
210 |
nm3 |
|