|
|
|
|
|
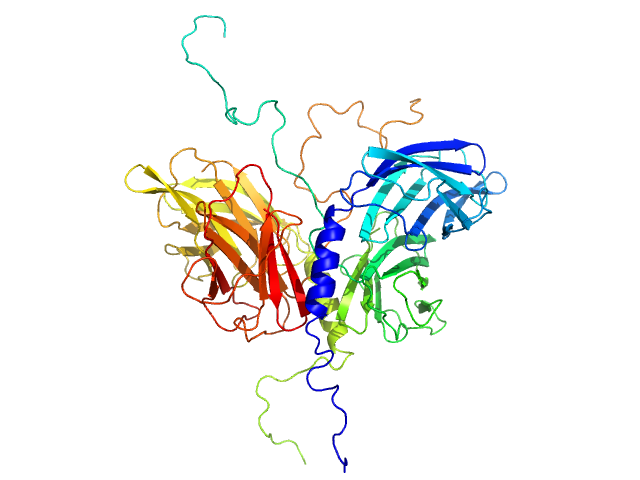
| Sample: |
Stator protein FlaG soluble domain dimer, 30 kDa Sulfolobus acidocaldarius protein
Conserved flagellar protein FlaF-I96Y soluble domain dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
25 mM citric acid/sodium citrate, 150mM NaCl, 3% Glycerol, pH: 3
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 10
|
The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility.
Nat Microbiol (2019)
Tsai CL, Tripp P, Sivabalasarma S, Zhang C, Rodriguez-Franco M, Wipfler RL, Chaudhury P, Banerjee A, Beeby M, Whitaker RJ, Tainer JA, Albers SV
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
90 |
nm3 |
|