|
|
|
|
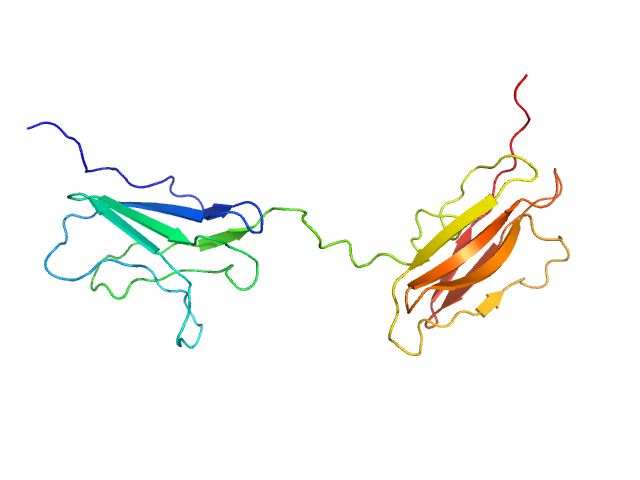
|
| Sample: |
Obscurin Ig domains 12/13 long monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 20
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
...Miller CJ, Wright NT
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
26 |
nm3 |
|