|
|
|
|
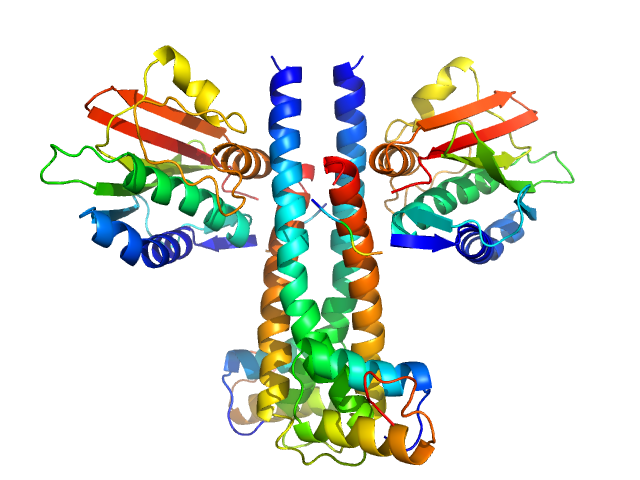
|
| Sample: |
Sporulation kinase A dimer, 51 kDa Bacillus subtilis protein
Sporulation inhibitor sda dimer, 11 kDa Bacillus subtilis protein
|
| Buffer: |
50mM Tris, 200mM NaCl, 150mM Imidazole, pH: 8.5
|
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Nov 16
|
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol 368(2):407-20 (2007)
Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, Guss JM, Trewhella J, Langley DB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
85 |
nm3 |
|