|
|
|
|
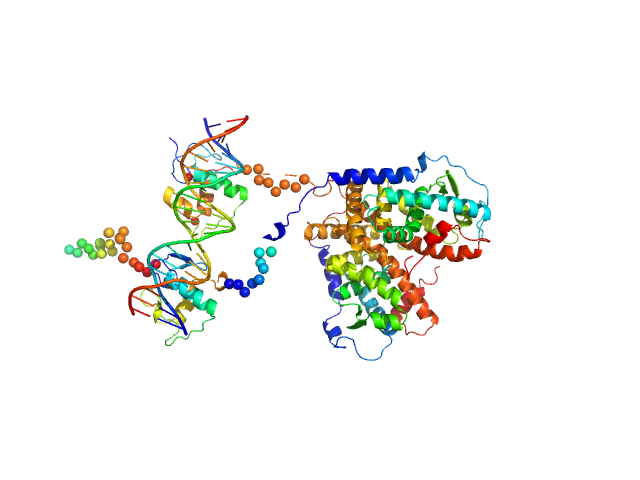
|
| Sample: |
Retinoic acid receptor alpha, RAR monomer, 41 kDa Mus musculus protein
Retinoic acid receptor RXR-alpha monomer, 38 kDa Mus musculus protein
...ment F11r DR5 monomer, 13 kDa DNA
|
| Buffer: |
...mM Tris, pH 8, 150 mM NaCl, 5% v/v glycerol, 1 mM CHAPS, 4 mM MgSO4, 1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at ...MBL P12, PETRA III on 2014 Jan 19
|
Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR
Nucleic Acids Research (2020)
...McEwen A, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries C, Svergun D, Rochel N
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
130 |
nm3 |
|